R still R still is the tool you want

We all know python popularity among DS practitioners has soared over the past few years, signaling both aspiring DS on the one hand and organizations on the other to favor python over R in a snowballing dynamic.
I’m writing this post to help turn the tide and let us all keep writing in the language we love and are most productive with. Feel free to join in on the effort by contributing to the “R advantages over python” repo and by joining the discussion on this post on Medium and my blog.
One popular way of demonstrating the rise of python is to plot the fraction of questions asked on stack overflow with the tag “pandas”, compared with “dplyr”:
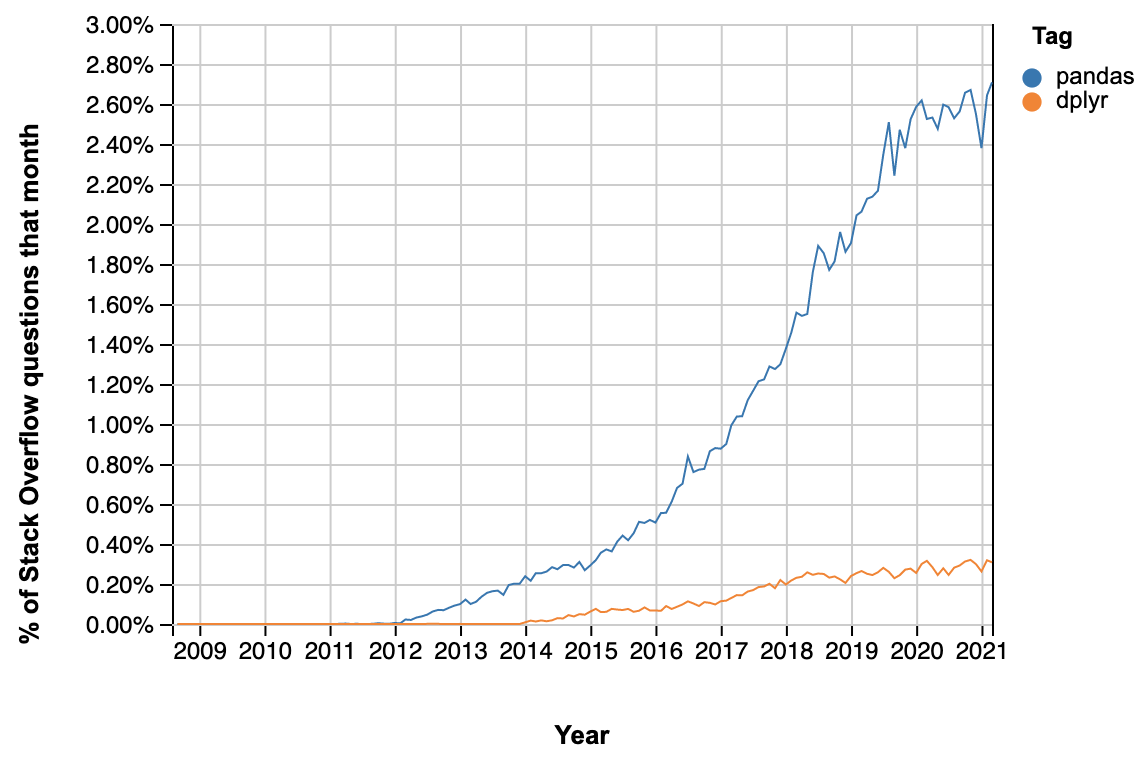
But there is also another story this graph is telling: All those new pandas users search stack overflow excessively because pandas is really unintelligible. Later in this post I’ll provide several examples of common operations that in dplyr are straightforward yet in pandas would require most of us to search stack overflow.
I’m a big believer in using the right tool for the job (I’ve been writing Scala Spark for the last 6 months for our Java based production environment). It has become common wisdom that for most data scientists data wrangling makes up most of the job.
Taken with permission from here
It follows that dplyr users enjoy a productivity boost most of the time compared with pandas users. R has the edge in a wide range of other areas too: available IDEs, package management, data constructs and many others.
Those advantages are so numerous that covering them all in a single post would be impractical. To that end I’ve started compiling all the advantages R has over python in a dedicated github repo. I plan on sharing them in small batches in a series of posts starting with this one.
When enumerating the different reasons I try to avoid the following:
- Too subjective comparisons. E.g. function indentation vs curly braces closure
- Issues that one can get used to after a while like python indexing (though the fact it starts from 0, or that object[0:2] returns only the first 2 elements still throws me off occasionally).
What do I hope to accomplish by this you’d might ask? I hope that:
- Organizations realize the value in working with R and choose to use it with/instead of python more often.
- As a result the python developer community becomes aware of the ways it can improve it’s DS offering and acts on them. python has already borrowed some great concepts from R (e.g. data frames, factor data type, pydatatable, ggplot) - it’s important it does so even more so we get to enjoy them when we have to work with python.
Now, I don’t argue R is preferable to python in every imaginable scenario. I just hope that becoming aware of the advantages R has to offer would encourage organizations to consider using it more often.
OK, so now for the burden of proof. Below I’ll give a few examples that show why in dplyr most operations are straightforward while in pandas they often require searching stack overflow. Again - this is just why dplyr is much easier to work with than pandas. For other advantages R has over python see the repo. We’ll start with a simple example: calculate the mean Sepal length within each species in the iris dataset.
Aggregation
We’ll start with a simple example: calculate the mean Sepal length within each species in the iris dataset.
In dplyr:
iris %>%
group_by(Species) %>%
summarise(mean_length = mean(Sepal.Length))A common way of doing the same in pandas would be using the agg method:
(
iris.groupby('Species').agg({'Sepal.Length':'mean'})
.rename({'Sepal.Length':'mean_length'}, axis = 1)
)We can see that pandas requires an additional rename call.
We can avoid the additional rename by passing a tuple to agg:
iris.groupby('Species').agg(mean_length = ('Sepal.Length', 'mean'))While this looks much closer to the dplyr syntax, it also highlights the fact there’s multiple ways of using the agg method - contrary to common wisdom that in R there are many ways to do the same thing while in python there’s only a single obvious way.
Now let’s say we’d like to use a weighted average (with sepal width as weights).
In dplyr we’d use the weighted mean function with an additional argument:
iris %>%
group_by(Species) %>%
summarize(weighted_mean_length = weighted.mean(Sepal.Length, Sepal.Width))Pretty straight forward. In fact, it’s so straight forward we can do the actual weighted mean calculation on the fly:
iris %>%
group_by(Species) %>%
summarize(weighted_mean_length = sum(Sepal.Length * Sepal.Width) / sum(Sepal.Width))In pandas it’s not so simple. One can’t just tweak the above examples. To come up with a pandas version I had to search stack overflow and based on this and this answers I got:
def weighted_mean(group):
d = {}
x = group['Sepal.Length']
w = group['Sepal.Width']
d['weighted_mean_length'] = (x * w).sum() / w.sum()
return pd.Series(d, index=['weighted_mean_length'])
sepal_length_to_width_ratio = (
iris
.groupby('Species')
.apply(weighted_mean)
)We can see that:
1. We have to define a custom function, and it can’t even work for general inputs
but rather has to have them hard coded.
2. The syntax is super cumbersome and requires searching stack overflow.
3. We need to use apply instead of the common agg method.
4. I’m pretty sure anyone not using the above code for more than a few weeks
would have to search stack overflow/his code base again to find the answer next
time he needs to do that calculation.
Window functions
Aggregation over a window
Let’s say we’d like to calculate the mean of sepal length within each species and append that to the original dataset (In SQL: SUM(Sepal.Length) OVER(partition by Species)) would be:
iris.assign(mean_sepal = lambda x: x.groupby('Species')['Sepal.Length'].transform(np.mean))We can see that this requires a dedicated method (transform), compared with dplyr which only requires adding a group_by:
iris %>%
group_by(Species) %>%
mutate(mean_sepal = mean(Sepal.Length))Now let’s say we’d like to do the same with a function that takes 2 argument variables. In dplyr it’s pretty straight forward and again, just a minor and intuitive tweak of the previous code:
iris %>%
group_by(Species) %>%
mutate(mean_sepal = weighted.mean(Sepal.Length, Sepal.Width))I wasn’t able to come up with a way to use a function with more than 1 input such as weighted mean in pandas.
Expanding windows
Now let’s say we’d like to calculate an expanding sum of Sepal.Length over increasing values of Sepal.Width within each species (in SQL: SUM(Sepal.Length) OVER(partition by Species ORDER BY Sepal.Width))
In dplyr it’s pretty straight forward:
iris %>%
arrange(Species, Sepal.Width) %>%
group_by(Species) %>%
mutate(expanding_sepal_sum = sapply(1:n(), function(x) sum(Sepal.Length[1:x])))Notice we don’t need to memorize any additional functions/methods. You find a solution using ubiquitous tools (e.g. sapply) and just plug it in the dplyr chain.
In pandas we’ll have to search stack overflow to come up with the expanding method:
(
iris.sort_values(['Species', 'Sepal.Width']).groupby('Species')
.expanding().agg({'Sepal.Length': 'sum'})
.rename({'Sepal.Length':'expanding_sepal_sum'}, axis = 1)
)Again, we need to use an additional rename call.
You’d might want to pass a tuple to agg like you’re used to in order to avoid the additional rename but for some reason the following syntax just wont work:
(
iris.sort_values(['Species', 'Sepal.Width']).groupby('Species')
.expanding().agg(expanding_sepal_sum = ('Sepal.Length', 'sum'))
)You could also avoid the additional rename by using the following eye sore:
(
iris.assign(expanding_sepal_sum = lambda x:x.sort_values(['Species', 'Sepal.Width'])
.groupby('Species').expanding().agg({'Sepal.Length': 'sum'})
.reset_index()['Sepal.Length'])
)Moving windows
Now let’s say we’d like to calculate a moving central window mean (in SQL: AVG(Sepal.Length) OVER(partition by Species ORDER BY Sepal.Width ROWS BETWEEN 2 PRECEDING AND 2 FOLLOWING))
As usual, in dplyr it’s pretty straightforward:
iris %>%
arrange(Species, Sepal.Width) %>%
group_by(Species) %>%
mutate(moving_mean_sepal_length = sapply(
1:n(),
function(x) mean(Sepal.Length[max(x - 2, 1):min(x + 2, n())])
))As in the other examples, all you have to do is find a solution using ubiquitous tools and plug it in the dplyr chain.
In pandas we’d have to look up the rolling method, read it’s documentation and come up with the following:
(
iris.sort_values(['Species', 'Sepal.Width']).groupby('Species')
.rolling(window = 5, center = True, min_periods = 1).agg({'Sepal.Length': 'mean'})
.rename({'Sepal.Length':'moving_mean_sepal_length'}, axis = 1)
)That’s it for this post. If you’re still not a convert watch out for more examples on the next post in this series. You can also check out more examples in the “R advantages over python” repo.